Publications
Research articles
-
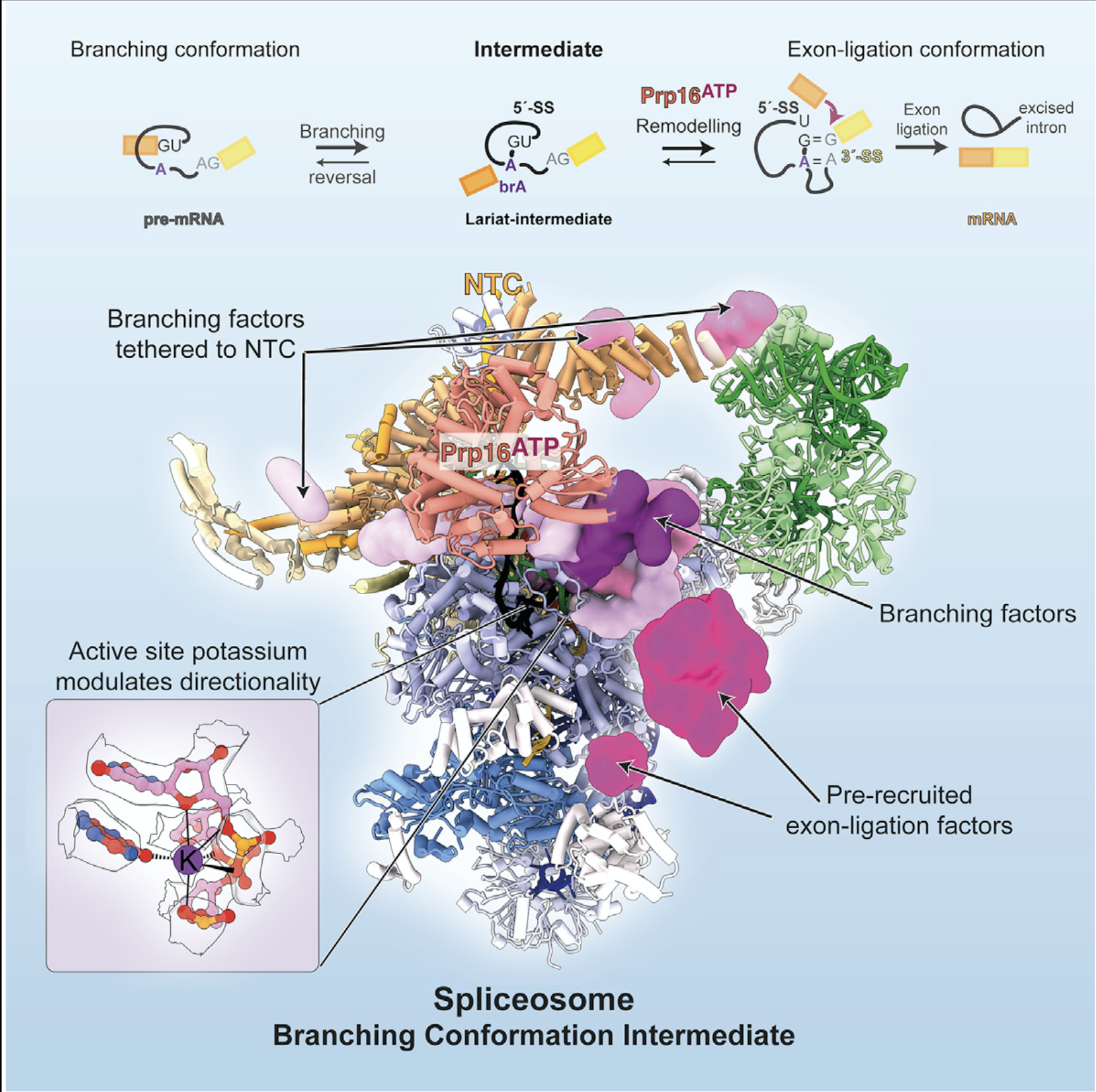
*Wilkinson, M. E.#, *Fica, Sebastian M.#, *Galej, W. P. & Nagai, K. Structural basis for conformational equilibrium of the catalytic spliceosome. Molecular Cell, 81,1-14 (2021), *equal contribution, #corresponding authors.
Genetic and biochemical studies suggested the two steps of pre-mRNA splicing are in thermodynamic equilibrium through two major conformations of the spliceosome during catalysis. Here, the structure of a new spliceosome intermediate between these two conformations reveals how binding of step-specific protein factors establishes equilibrium of the spliceosome.
Featured in a Q&A in Molecular Cell.
-
Strittmatter, L. M., Capitanchick, C., Newman, A. J., Hallegger, M., Norman, C. M., Fica, Sebastian M.#, Oubridge, C., Luscombe, N.M.#, Ule, J.#, Nagai, K. PsiCLIP reveals dynamic RNA binding by DEAH-box helicases before and after exon ligation. Nature Communications, 12, 1488 (2021). #corresponding authors.
ATP-dependent helicases remodel the spliceosome and proofread splice site recognition. Here the Nagai and Ule labs developed Purified Spliceosome iCLIP (psiCLIP) to probe protein-RNA interactions in defined spliceosome complexes and reveal how the helicases Prp16 and Prp22 promote correct mRNA synthesis through dynamic binding on their RNA substrates.
Featured in an MRC LMB Insight on Research.
-
Fica, Sebastian M.#, Oubridge, C., Wilkinson, M. E., Newman, A. J., Nagai, K.# (2019) A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science,
363, 710-714, #corresponding authors.
The structure of a human post-catalytic spliceosome showed how mammalian spliceosomes co-opted additional protein factors to stabilize 3'-splice site docking. We discovered a metazoan-specific splicing factor important for mRNA synthesis. This factor is a tumor suppressor, which potentially implicates regulation of splicing catalysis in cancer pathology.
Featured in an MRC LMB Insight on Research.
-
Wilkinson, M. E., Fica Sebastian M., Galej, W. P., Norman, C. M., Newman, A. J., Nagai, K. (2017) Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection. Science, 358, 1283-1288.
This structure of a yeast post-catalytic spliceosome revealed the mechanism for 3'-splice site docking at the active site.
Featured in an MRC LMB Insight on Research.
-
Fica, Sebastian M.#, Oubridge, C., Galej, W. P., Wilkinson, M. E., Bai, X.-C., Newman, A. J., & Nagai, K.# (2017). Structure of a spliceosome remodelled for exon ligation. Nature, 542,
377-380, #corresponding authors.
This work elucidated the spliceosome conformation competent to perform the second catalytic step of splicing for mRNA synthesis.
-
Galej, W. P., Wilkinson, M. E., Fica, Sebastian M., Oubridge, C., Newman, A. J., & Nagai, K. (2016). Cryo-EM structure of the spliceosome immediately after branching. Nature, 537,
197-201.
This work presented one of the first two structures of a yeast catalytic spliceosome and revealed the structure of the active site and the role of branching factors.
-
*Fica, Sebastian M., *Mefford, M. A., Piccirilli, J. A., & Staley, J. P. (2014). Evidence for a group II intron-like catalytic triplex in the spliceosome. Nature Structural & Molecular Biology,
21, 464-471, *equal contribution.
This study demonstrated that the active site of the spliceosome forms an RNA triple helix, which is essential for proper positioning of the two active site catalytic metals.
-
*Fica, Sebastian M., *Tuttle, N., Novak, T., Li, N.-S., Lu, J., Koodathingal, P., Dai, Q., Staley, J. P., Piccirilli, J. A. (2013). RNA catalyses nuclear pre-mRNA splicing. Nature,
503, 229-234. *equal contribution.
This study presented definitive functional evidence that the spliceosome active site is composed of RNA, while proteins stabilise and organise the RNA-based active site but do not play a direct catalytic role.
-
Schellenberg, M. J., Wu, T., Ritchie, D. B., Fica, Sebastian M., Staley, J. P., Atta, K. A., LaPointe, P. L., MacMillan, A. M. (2013). A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation. Nature Structural & Molecular Biology, 20, 728-734.
Review articles
-
Fica, Sebastian M.# (2020). Cryo-EM snapshots of the human spliceosome reveal structural adaptions for splicing regulation. Current Opinion in Structural Biology, 65, 139-148. #corresponding
author.
This review provides a comprehensive summary of key structures of the human spliceosomes and conceptualizes the roles of human-specific splicing factors discovered in recent structures. These factors are likely to modulate splice site choice and alternative splicing.
-
Fica, Sebastian M.#, & Nagai, K.# (2017). Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nature Structural & Molecular Biology,
24, 791-799, #corresponding authors.
This review provided an overview of the initial spliceosome structures obtained by electron microscopy and placed their findings in the context of previous genetic and biochemical studies.
-
Nguyen, T. H. D., Galej, W. P., Fica, Sebastian M., Lin, P.-C., Newman, A. J., & Nagai, K. (2015). CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast. Current Opinion in Structural Biology, 36, 48-57.
Book chapters
-
Fica, Sebastian M., Small, E.C., Mefford, M., and Staley, J. P., Mechanistic Insights into Mammalian Pre-mRNA Splicing, Posttranscriptional Gene Regulation: RNA Processing in Eukaryotes, Jane Wu Ed, First Edition, Wiley
(2013).