Research
Native complex purification
RNA Biochemistry
Electron Microscopy
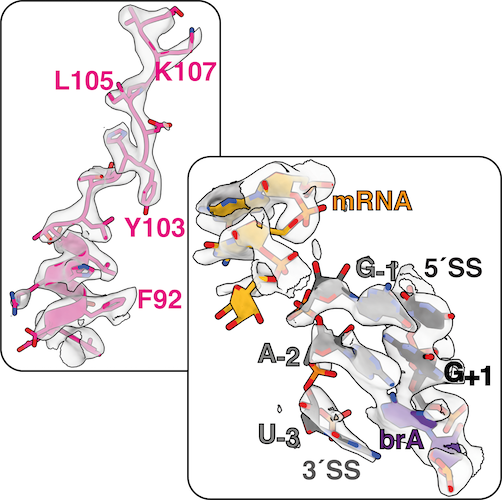
Cryo-EM Maps & Atomic Models
Splicing Catalysis
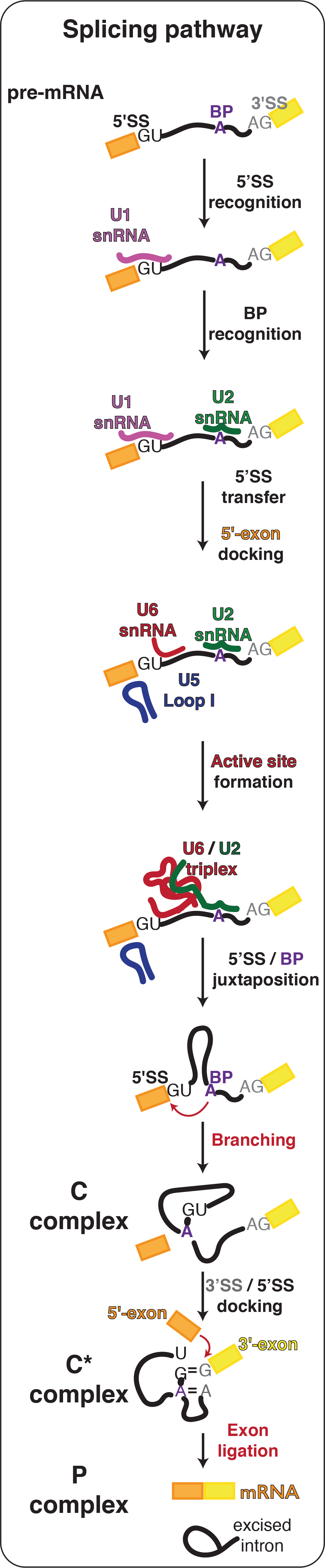
The active site of the spliceosome is formed by the U2 and U6 snRNAs, which form a triple helix that allows U6 snRNA to bind two catalytic Mg2+ ions. The 5´-splice site (5´SS) is positioned at the catalytic metal ions by pairing between the 5´ end of the intron and the U6 snRNA and between the 5´-exon and U5 snRNA. During spliceosome activation, the branch adenosine (brA) is docked into the active site, where its 2´-hydroxyl attacks the 5´SS during branching, producing the free 5´-exon and a lariat intron-3´exon intermediate. The brA then rotates out of the active site, alowing the the 3´SS to dock instead for exon ligation. The 3´-SS is positioned at the catalytic metal ions by non-Watson-Crick basepairing between the last intron nucleotide G and the first intron nucleotide G as well as between the penultimate intron nucleotide A and the brA. This configuration allows the 3´-hydroxyl group of the 5´-exon to attack the 3´SS, linking the 5´- and 3´-exons into mRNA during exon ligation.
The structure of the active site remains essentially the same during both catalytic steps and recent work with Max Wilkinson in the Nagai lab suggests that a monovalent metal ion is also necessary to stabilise the catalytic configuration of the active site. This third metal ion - K1 - appears to bind to the triplex only during the final stages of activation and we are keen to investigate its potential role in modulating the equilibrium between the branching and exon ligation conformations of the active site.
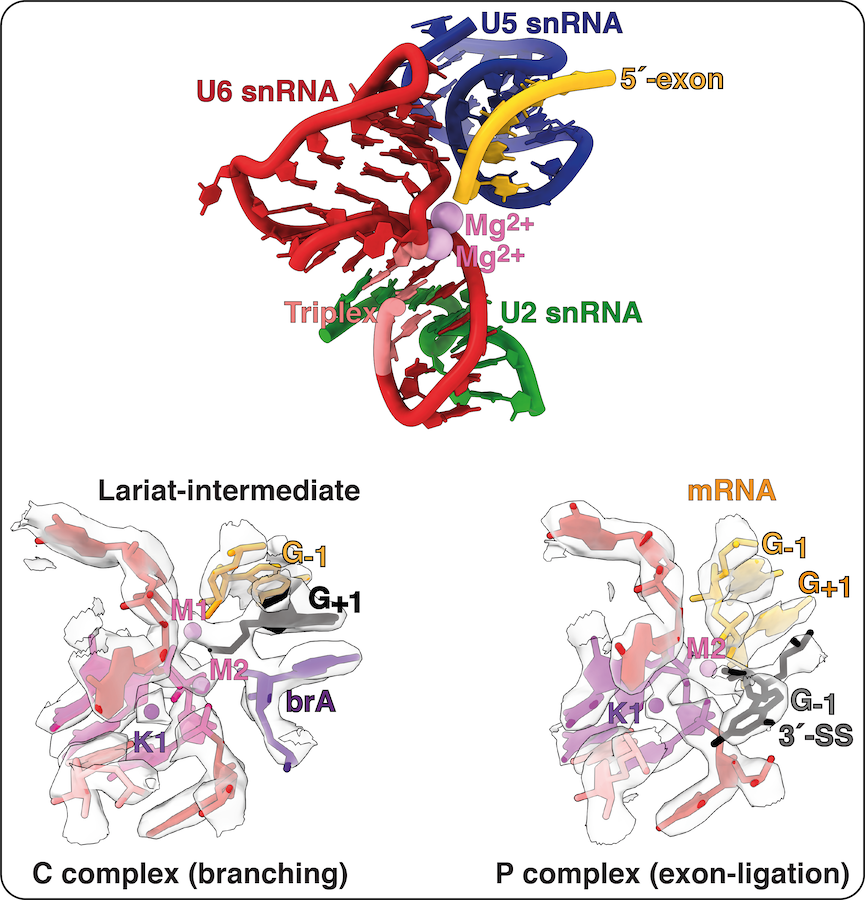
Human splicing factors
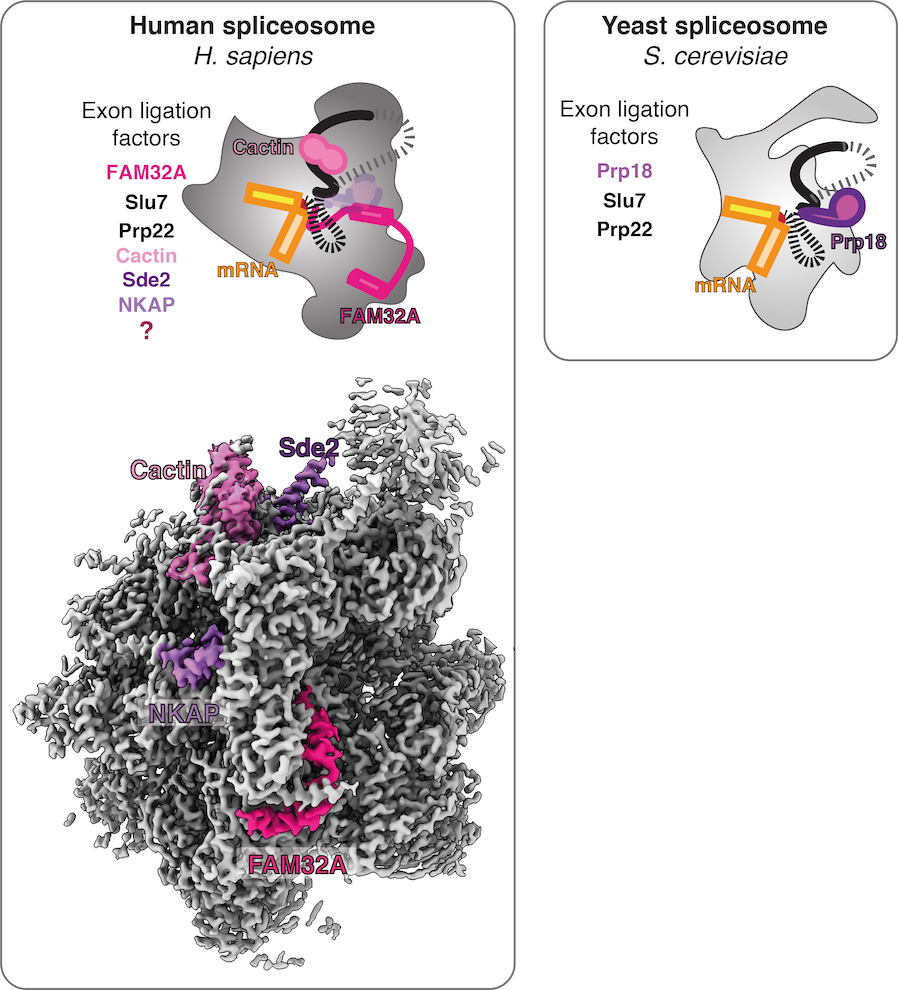
Although the active site is conserved between yeast and humans, the mechanisms for remodelling and the specific roles of trans-acting factors have been barely studied in mammals. Besides the basic protein factors observed in yeast, many additional trans-acting factors of yet unknown role associate with the human spliceosome during the catalytic phase (Fica, 2020). Indeed, we recently discovered that mammalian spliceosomes employ additional exon ligation factors not seen in yeast (Fica et al., 2019), highlighting the catalytic stage as an unexplored target for regulation of splicing. While in yeast Prp18 is constitutively bound to C* and P spliceosomes during exon ligation, in humans Prp18 is partially substituted by FAM32A and appears to bind the C* complex in a transcript-specific manner (Fica et al., 2019; S. Fica and K. Nagai, manuscript in preparation). Together these observations suggest that splicing of specific pre-mRNAs may rely on different complements of exon ligation factors.
More boradly, the mechanisms underlying correct splice site choice during catalysis are unclear. While alternative splicing, splice site choice, and trans-acting factors have been studied extensively at the pre-catalytic stage, before the C complex, the final splice site choice must be made during the C to C* transition when the spliceosome chooses the 3′-SS for exon ligation, as this is when the mRNA is synthesized. Thus, the many trans-acting factors that bind the human catalytic spliceosome could influence splice site selection and alternative splicing at the C* stage, but their role in fidelity has not been studied.
We aim to investigate at the molecular level how mammalian exon ligation factors bind to and remodel the catayltic spliceosome to promote splicing of specific pre-mRNAs.
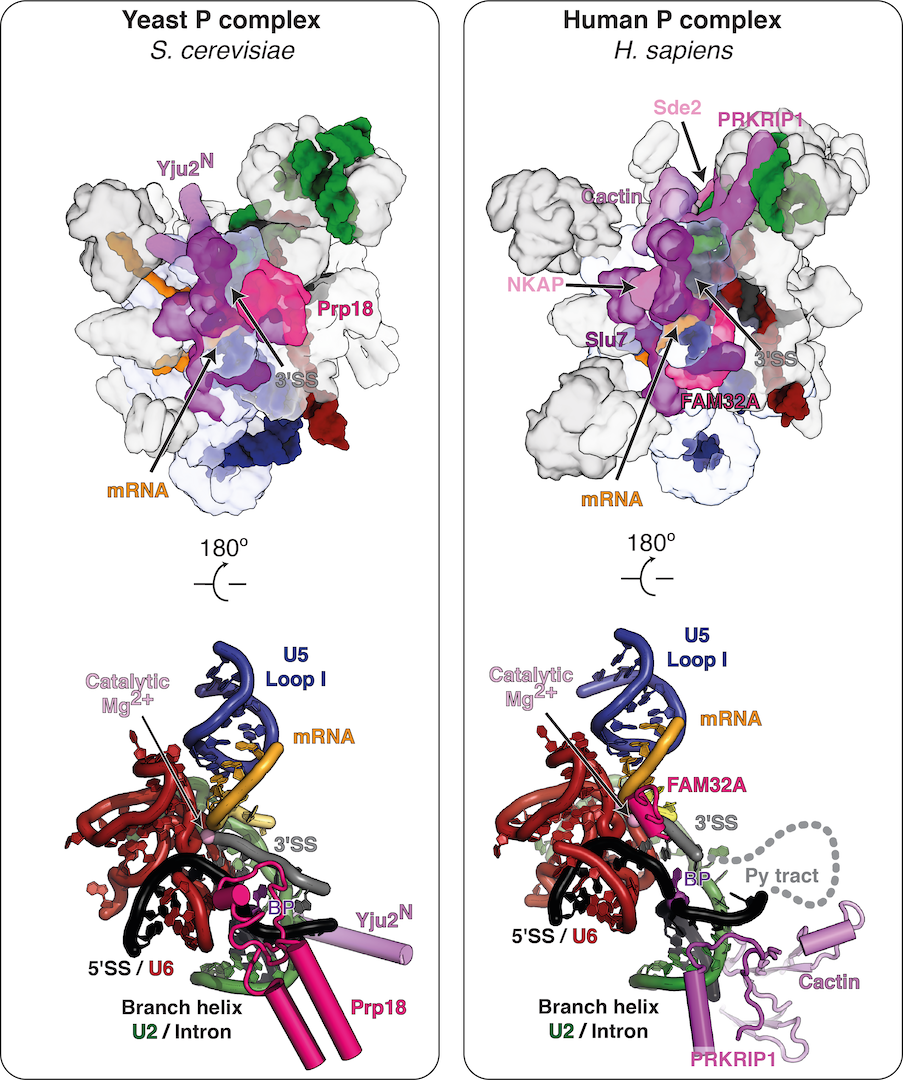
Structural proteomics
In humans, many more factors associate at the catalytic stage compared to yeast (Fica et al., 2019; Fica, 2020). Until recently, it was unknown how these additional factors bind the spliceosome and what role they play during catalysis. By solving high-resolution structures of human spliceoosmes at the exon-ligation stage, we were able to identify several of these factors from density alone. Thus we were able to discern the stoichimetric binding of specific factors by interpreting our cryo-EM maps, rather than by traditional biochemistry or mass spectrometry.
We found that some human exon ligation factors (e.g. Prp18) may only engage the spliceosome assembled on specific transcripts (S. Fica and K. Nagai, manuscript in preparation). Other exon ligation factors (e.g. Cactin) have the potential to influence docking of the 3′-SS. Indeed, Cactin affects splicing of specific introns in fisison yeast and in humans. These observations suggest that specific transcript sequences could determine the complement of exon ligation factors that are necessary for splicing of specific pre-mRNAs. Such a model would explain why mass spectrometric studies of the human spliceosome have consistently revealed numerous substoichiometric factors that have not yet been observed in any spliceosome structures, as all published structures have been assembled on Minx pre-mRNA and some factors may require other specific transcripts to engage the spliceosome core stably and be visualized. We thus aim to purify human spliceosomes assembled on diverse pre-mRNAs from different tissue types and developmental states, with the goal of using structural biology to identify where all human splicing factors bind. Structures of native complexes from diverse tissues could also lead to the discovery of novel human splicing factors that act during the catalytic stage.
Spliceosome dynamics
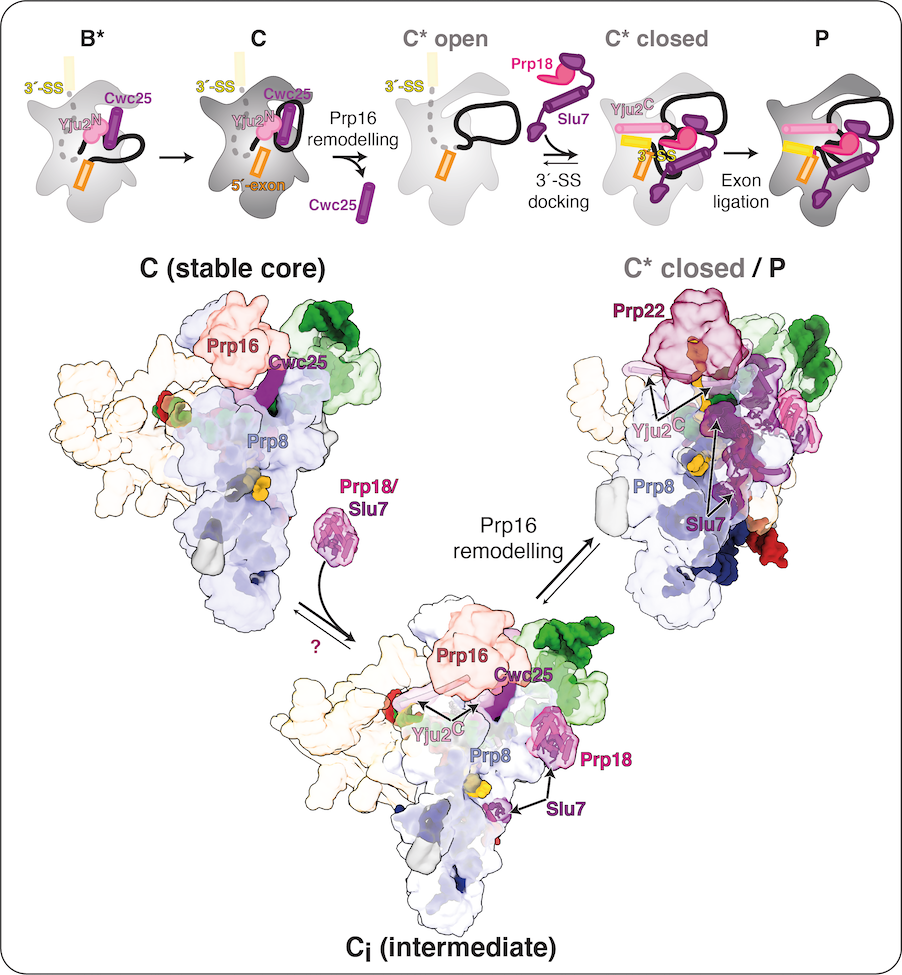
While a single active site catalyses both splicing steps (Fica et al., 2013; Galej et al., 2016; Wilkinson et al., 2017), a structural rearrangement allows the 3′-SS to replace the branch helix in the active site during exon ligation. This rearrangement is promoted by the ATPase Prp16, which governs the equilibrium between the branching (B*/C) and exon ligation (C*/P) conformations of the catalytic spliceosome. Prp16 promotes dissociation of the branching factor Cwc25 to allow binding of exon-ligation factors. Working with Max Wilkinson and Wojtek Galej in the Nagai lab, we identified a novel C-complex intermediate (Ci) that elucidated the molecular basis for this equilibrium. The exon-ligation factors Prp18 and Slu7 are already bound in Ci, before ATP hydrolysis by Prp16 can destabilise the branching conformation. Biochemical assays suggest these pre-bound factors prime C complex for conversion to C* by Prp16. Indeed, some branching factors such as Yju2 remain bound to the spliceosome during exon ligation although their binding is remodelled by Prp16 action.
Our work thus suggests that dynamic binding of step-specific factors stabilizes either the branching or exon-ligation conformation and allows the spliceosome to toggle between these two states during catalysis. We are keen to explore this equilibrium in the human spliceosome, where many more step-specific factors engage during the catalytic stage. The binding of these factors could modulate conformational equilbrium during catalysis in a manner that affects more broadly proofreading and splice site selection, particularly during exon ligation.